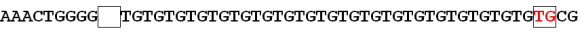Out-of-India - From Theory to Truth: Part 2
Re: Out-of-India - From Theory to Truth: Part 2
Whole e-book is better doc. You can separately blogify it also ... No harm done. This Asko Parpola chap has come out with something new now btw... not sure if you have seen it. Will get the link and post it here.
Found it:
https://www.academia.edu/18861957/Parpo ... _7_2_14-15
Found it:
https://www.academia.edu/18861957/Parpo ... _7_2_14-15
Re: Out-of-India - From Theory to Truth: Part 2
Link to video"You Aryans have come from outside." - Mallikarjun Kharge's intervention
[youtube]QWOi6lHxIJ4?t=97[/youtube]
Published on Nov 26, 2015
Rajnath Singh: If Sardar Patel is the uniting force, BR Ambedkar is the binding force. Dr. Ambedkar introduced the concept of reservation for equality, he said it's a socio-political necessity."
Rajnath says our founding fathers believed that the Preamble is the soul of the Constitution and no changes should be made to that. 'Socialist' aur 'Secular' in do shabdon ko insert kia gaya, mai nahi kehta hoon mai use nahi maanta, jo ho gaya so ho gaya.
Mallikarjun Kharge, leader of the opposition in the lower house objects to Rajnath's statement saying Dr Ambedkar wanted insert the words socialist and secular in the preamble but was not allowed to. He also said Rajnath's contention that Dr Ambedkar wanted to leave the country was wrong. "Dr Ambedkar never wanted to leave the country. He was part of the country. You Aryans have come from outside."
Here, congress leader in Parliament is talking about Ambedkar and endorsing Aryan Invasion Theory. Ambedkar himself rejected AIT, but congress leader is pushing AIT in Parliament.
Link“Ambedkar did not endorse Aryan-Dravidian theory”
B. R. Ambedkar never propounded Brahmin/non Brahmin binary and also never endorsed the Aryan-Dravidian Theory of race. “Ambedkar was indeed much more intuitive on these issues; he rejected the ‘Aryan-non-Aryan theory' as a historical justification and asserted that caste was vastly different from race”, said Kudiyarasan, a writer based in Chennai.
Addressing a seminar organised by Viduthalai Chiruthaigal Katchi on Sunday to mark the 121{+s}{+t}birth anniversary of Dr. Ambedkar, he said that he carefully recorded that the enemy was not ‘Brahmans' but ‘Brahmanism', which he harshly attacked but defined not in terms of a specific group but as the negation of the values of freedom, liberty, equality and fraternity. His theory was based on the binary touchable/untouchable based on theories of caste.
Mr. Kudiyarasu further said that the Backward Classes leader and leaders of Dravidian movement were comfortable and receptive towards Dr. Ambedkar and his ideas but at the same time they never recognised regional Dalit leaders like Iyothee Thass Pandithar, M. C. Rajah, Rettamalai Srinivasan or N. Sivaraj.
The works of Rajah, Rettamalai Srinivasan, Sivaraj and Meenambal Sivaraj were suppressed by the movements that were propagating Dravidian and non-Brahmin inclusiveness. “It was the Dalit woman icon Meenambal Sivaraj, who was also the head of the South Indian Scheduled Caste Federation, who gave the title ‘Periyar' (Great One) to E. V. Ramasamy Naicker but facts like these were covered up and did not reach the popular discourse”, he pointed out.
T. Lajapathi Roy, Advocate, Madras High Court, in his talk stated that Dr. Ambedkar was a man of sharp ideas.
LinkKnowing Ambedkar
Sunday, 14 April 2013 | CHANDRA BHAN PRASAD | in Bignames
7
Time we learnt true teachings of Baba Saheb
Today is Dr BR Ambedkar’s Birth Anniversary and the best way to remember him is to discuss his ideas and also examine what his followers thought of those ideas.
When I was in Class VIII, the Dalits in my village formed a sports club. The club was named Dr Ambedkar Kirti Club. Incidentally, Dr Ambedkar Kirti Club was given by Munoo Rai an upper caste of the village. The fact that we had a club named after Dr Ambedkar meant that the elder Dalits in the village would often talk about him and his ideas. In fact, the very next year, some Dalits even raised the question of launching a Dalit Panthar (a social organisation committed obtaining social and economic justice for all) in our village.
I had thus, in my very childhood, an introductory lesson on Ambedkar. Three images formed in the mind when I thought of him:
A man who drafted the India’s Constitution
Author of Arya-non-Arya thesis
A person who re-igniter the dhamma chakra
A few years back, I found out, much to my chagrin, that I had been taught a wrong lesson. The lesson we had learnt was that we (the Dalits) the non-Aryans had been invaded, tormented and enslaved by the caste-Hindu upper caste who were Aryans.
Ask any Dalit today (a Dalit with doctorate on Dr Ambedkar and/or a Dalit school dropout) about Dr Ambedkar’s Aryan invasion theory and most will tell you that Baba Saheb was right when he talked about the Aryan invasion theory.
But how much truth is there? In his book Who Were the Shudras Vol 7, Dr Ambedkar dismisses the Aryan race with absolute contempt. On page 85, he concludes by saying (while dismissing the Aryan race theory) that:
The Vedas do not know any race referred to as the Aryan
There is no evidence in the Vedas of any invasion of India by the Aryan race and its having conquered the Dasas and Dasyus who were supposed to be natives of India
There is no evidence to show that the distinction between Aryans, Dasas, and Dasyus was a racial distinction
The Vedas do not support the contention that the Aryans were different in colour from the Dasas and Dasyus
In the same book (page 86), Dr Ambedkar writes:
“Enough has been said to show how leaky is the Aryan theory expounded by western scholars and accepted by Brahmins. Yet, the theory has such a hold on the people that what has been said against it may mean no more than scotching it. Like the snake, it must be killed.”
On page 100, Dr Ambedkar writes: “In the face of the discovery of new facts set out in this chapter, the theory can no longer stand and must be thrown on the scrap heap.”
It is a real puzzle that Dr Ambedkar’s followers believe in the Aryan race theory which he himself rejected. To him, Aryan is a linguistic term and not a term for race. Worse still, Dr Ambedkar’s followers believe that it was Dr Ambedkar himself who propounded the Aryan race theory!
When late Kanshi Ram came up with his theory of Bahujans, Dalits in the north believed the Dalits-OBCs are always the victims and the enemy is always the upper caste.
Dr Ambedkar had no such ambiguity. The book Who Were the Shudra is in fact two books put together as one. The first part is titled Who Were the Shudra and dedicated to Mahatma Jyotirao Phule. In his dedication to Phule, Dr Ambedkar writes: “The greatest shudra of modern India who made the lower classes of Hindus conscious of their slavery to the higher classes and who preached the gospel that for India social democracy was more vital than independence from foreign rule.”
The second part of the book is titled The Untouchables. This part of the book is dedicated to three Dalits saints — Nandanar (Tamil Nadu), Guru Ravidass (Uttar Pradesh) and Chokhamela (Maharashtra). It is one thing to propound or believe in a theory or ideology and it is quite other thing to ascribe that theory or ideology to some one who never propounded it.
If we have to learn from Dr Ambedkar, the first step would be to unlearn what has been stored in the name of Dr Ambedkar. He never fabricated social categories and he would have been the first person to talk about what the reality is.
Re: Out-of-India - From Theory to Truth: Part 2
Says page unavailable. It seem the response draft didn't make it through the admins?shiv wrote:DoneEswarPrakash wrote:Could some knowledge person please give a good rebuttal to https://www.saddahaq.com/who-are-we-4471 because I don't have the capability to do it myself.
https://www.saddahaq.com/storydraft/f5f ... 667/rev/39
There's no other way to comment. No options to vote down, dislike or report. Only favorite, vote up, like and share buttons.
Quite rich that ... and lots of lies in the article.
But I think this topic is so old and discussed so many times that all arguments and counter arguments are there on net already. Not to bother too much on one article.
Re: Out-of-India - From Theory to Truth: Part 2
^^^the article is important as it exposes Mohan Guruswamy's shoddy understanding of history!
This man is so worried about ultra-nationalism that he is willing to ignore the exta-nationals invading the very identity of Indians.
The issue can be ignored at great peril given educated folks like Kharge and Gurusswamy endorse the AIT nonsense.
This man is so worried about ultra-nationalism that he is willing to ignore the exta-nationals invading the very identity of Indians.
The issue can be ignored at great peril given educated folks like Kharge and Gurusswamy endorse the AIT nonsense.
Re: Out-of-India - From Theory to Truth: Part 2
Are Kharge and Guruswamy Dalits? What is their religion does anyone know?Pulikeshi wrote:^^^the article is important as it exposes Mohan Guruswamy's shoddy understanding of history!
This man is so worried about ultra-nationalism that he is willing to ignore the exta-nationals invading the very identity of Indians.
The issue can be ignored at great peril given educated folks like Kharge and Gurusswamy endorse the AIT nonsense.
Re: Out-of-India - From Theory to Truth: Part 2
Kharge has the reputation of being anti-forward caste. Someone had told me this - but his remarks in parliament confirmed to me that he has a warped belief in his mind that echoes the racist views that the British made up at the turn of the century and was taught to Indians - a view that is now espoused by evangelists.peter wrote:Are Kharge and Guruswamy Dalits? What is their religion does anyone know?Pulikeshi wrote:^^^the article is important as it exposes Mohan Guruswamy's shoddy understanding of history!
This man is so worried about ultra-nationalism that he is willing to ignore the exta-nationals invading the very identity of Indians.
The issue can be ignored at great peril given educated folks like Kharge and Gurusswamy endorse the AIT nonsense.
The view is that North Indians are descended from whiteys and south Indian "dravidians" are descended from inferior blackies - and it is the mixture of superior whiteys with inferior blackies that cause the "degeneration" of Aryans in India. This view lasted till the Church needed more recruits. Now evangelists are saying that the "Inferior Dravidians" need to be made superior again by conversion to egalitarian faith
The term Dravidian itself is utter bullshit in the way it was conceived and is as bad the way "Aryan" was conceived by Europeans in order to make their own history older and better than the Semitic history that was discovered in the Levant region - a discovery that caused great heartburn in 18th century Europe where people could not conceive that Assyria had a history older than the Bible
Re: Out-of-India - From Theory to Truth: Part 2
The topic of DNA based genealogy mapping comes up in this thread many times. So, I was trying to understand this concept recently. Here is noob understanding so far:
Mainly, there seem to be two markers in DNA testing:
DNA Marker 1: Y Chromosome which is supposedly passed on from Father to son(not to daughter).
DNA Marker 2: Mitochondrial chromosome passed on by the mother to all her children(son & daughter).
That means, it is not possible to find the father of a daughter from genetic DNA testing because the daughter does not have the Y chromosome of the father. And women's DNA seems to be spread wider because all the kids inherit it. (Now, that itself raises some doubt in my mind about this field. I think all kids inherit the qualities from both parents and if the DNA is not able to study this aspect then it means the field has many shortcomings. The world is filled with daughters who look like their fathers).
So, lets say a person has 4 grandparents. Then, one can find the paternal grandfather using Y chromosome. Maternal grandmother and paternal grandmother both have contributed mitochondria thing. So, there is some chance of finding it, perhaps. But, one can't find the maternal grandfather with this DNA thing. Now, out of 4 grandparents: 1 can be found with certainty. 2 can be found with good probability. 1 can't be found.
But, as we go back in time, this becomes less and less effective. Because the number of ancestors keeps rising at each generation.
Lets say that a new generation of kids are born every 30 years. So, for a person in 2015:
1 person - yr 2015 CE
2(2^1))parents - yr 1985 CE
4(2^2) Grandparents - yr 1955 CE
8(2^3))Great Grandparents - yr 1925 CE
16(2^4) 4th generation ancestors - yr 1895 CE.
32(2^5) 5th generation ancestors - yr 1865 CE.
64(2^6) 6th generation ancestors - yr 1835 CE.
128(2^7) 7th generation ancestors - yr 1805 CE.
256(2^8) 8th generation ancestors - yr 1775 CE.
512(2^9) 9th generation ancestors - yr 1745 CE.
1024(2^10) 10th generation ancestors - yr 1715 CE.
2048(2^11) 11th generation ancestors - yr 1685 CE.
4096(2^12) 12th generation ancestors - yr 1655 CE.
8192(2^13) 13th generation ancestors - yr 1625 CE.
16,384(2^14) 14th generation ancestors - yr 1595 CE.
32,768(2^15) 15th generation ancestors - yr 1565 CE.
65,536(2^16) 16th generation ancestors - yr 1535 CE.
1,31,072(2^17) 17th generation ancestors - yr 1505 CE. 1 lakh
2,62,144(2^18) 18th generation ancestors - yr 1475 CE.
5,24,288(2^19) 19th generation ancestors - yr 1445 CE.
10,48,576(2^20) 20th generation ancestors - yr 1415 CE.
20,97,152(2^21) 21th generation ancestors - yr 1385 CE.
41,94,304(2^22) 22th generation ancestors - yr 1355 CE.
83,88,608(2^23) 23th generation ancestors - yr 1325 CE.
1,67,77,216(2^24) 24th generation ancestors - yr 1295 CE. 1 crore
3,35,54,432(2^25) 25th generation ancestors - yr 1265 CE.
6,71,08,864(2^26) 26th generation ancestors - yr 1235 CE.
13,42,17,728(2^27) 27th generation ancestors - yr 1205 CE.
26,84,35,456(2^28) 28th generation ancestors - yr 1175 CE. 25 crores
53,68,70,912(2^29) 29th generation ancestors - yr 1145 CE. 50 crores
107,37,41,824(2^30) 30th generation ancestors - yr 1115 CE. 107.375 crores = 1 Billion
214.7 crore = 2 Billion (2^31) 31th generation ancestors - yr 1085 CE.
429.4 crore = 4 Billion (2^32) 32th generation ancestors - yr 1055 CE.
858.8 crore = 8 Billion (2^33) 33th generation ancestors - yr 1025 CE.
1717.6 crore = 16 Billion (2^34) 34th generation ancestors - yr 995 CE.
3435.2 crore = 32 Billion (2^35) 35th generation ancestors - yr 965 CE.
6870.4 crore = 64 Billion (2^36) 36th generation ancestors - yr 935 CE.
13740.8 crore = 128 Billion (2^37) 37th generation ancestors - yr 905 CE.
27481.6 crore = 256 Billion (2^38) 38th generation ancestors - yr 875 CE.
54963.2 crore = 512 Billion (2^39) 39th generation ancestors - yr 845 CE. 500 Billion
109926.4 crore = 1024 Billion (2^40) 40th generation ancestors - yr 815 CE. 1000 Billion
In 815 CE, 1000 Billion ancestors of a single person in 2015 CE. And it keeps doubling because every person has 2 parents and each of those parents has 2 more parents and so on. The numbers are so huge that I don't know how it adds up. To give an idea, the present population of the world is 7 Billion. It seems as If I am missing something but I don't know what I am missing. If I am making some mistake please point out.
Broadly, it seems that the number of ancestors would be too large for even a single person as we go back in time and there seems to a total mixing making everyone relative of each other. So, it seems funny that people talk about AIT or something else based on these markers which become less probabilistic and totally mixed up as we go back in time. Only one ancestor can be found with certainty based on Y chromosome using DNA testing. Mitochondria is passed by every female ancestor, so given the huge number of ancestors , one can be sure that all sorts of Mitochondria will be there.
Mainly, there seem to be two markers in DNA testing:
DNA Marker 1: Y Chromosome which is supposedly passed on from Father to son(not to daughter).
DNA Marker 2: Mitochondrial chromosome passed on by the mother to all her children(son & daughter).
That means, it is not possible to find the father of a daughter from genetic DNA testing because the daughter does not have the Y chromosome of the father. And women's DNA seems to be spread wider because all the kids inherit it. (Now, that itself raises some doubt in my mind about this field. I think all kids inherit the qualities from both parents and if the DNA is not able to study this aspect then it means the field has many shortcomings. The world is filled with daughters who look like their fathers).
So, lets say a person has 4 grandparents. Then, one can find the paternal grandfather using Y chromosome. Maternal grandmother and paternal grandmother both have contributed mitochondria thing. So, there is some chance of finding it, perhaps. But, one can't find the maternal grandfather with this DNA thing. Now, out of 4 grandparents: 1 can be found with certainty. 2 can be found with good probability. 1 can't be found.
But, as we go back in time, this becomes less and less effective. Because the number of ancestors keeps rising at each generation.
Lets say that a new generation of kids are born every 30 years. So, for a person in 2015:
1 person - yr 2015 CE
2(2^1))parents - yr 1985 CE
4(2^2) Grandparents - yr 1955 CE
8(2^3))Great Grandparents - yr 1925 CE
16(2^4) 4th generation ancestors - yr 1895 CE.
32(2^5) 5th generation ancestors - yr 1865 CE.
64(2^6) 6th generation ancestors - yr 1835 CE.
128(2^7) 7th generation ancestors - yr 1805 CE.
256(2^8) 8th generation ancestors - yr 1775 CE.
512(2^9) 9th generation ancestors - yr 1745 CE.
1024(2^10) 10th generation ancestors - yr 1715 CE.
2048(2^11) 11th generation ancestors - yr 1685 CE.
4096(2^12) 12th generation ancestors - yr 1655 CE.
8192(2^13) 13th generation ancestors - yr 1625 CE.
16,384(2^14) 14th generation ancestors - yr 1595 CE.
32,768(2^15) 15th generation ancestors - yr 1565 CE.
65,536(2^16) 16th generation ancestors - yr 1535 CE.
1,31,072(2^17) 17th generation ancestors - yr 1505 CE. 1 lakh
2,62,144(2^18) 18th generation ancestors - yr 1475 CE.
5,24,288(2^19) 19th generation ancestors - yr 1445 CE.
10,48,576(2^20) 20th generation ancestors - yr 1415 CE.
20,97,152(2^21) 21th generation ancestors - yr 1385 CE.
41,94,304(2^22) 22th generation ancestors - yr 1355 CE.
83,88,608(2^23) 23th generation ancestors - yr 1325 CE.
1,67,77,216(2^24) 24th generation ancestors - yr 1295 CE. 1 crore
3,35,54,432(2^25) 25th generation ancestors - yr 1265 CE.
6,71,08,864(2^26) 26th generation ancestors - yr 1235 CE.
13,42,17,728(2^27) 27th generation ancestors - yr 1205 CE.
26,84,35,456(2^28) 28th generation ancestors - yr 1175 CE. 25 crores
53,68,70,912(2^29) 29th generation ancestors - yr 1145 CE. 50 crores
107,37,41,824(2^30) 30th generation ancestors - yr 1115 CE. 107.375 crores = 1 Billion
214.7 crore = 2 Billion (2^31) 31th generation ancestors - yr 1085 CE.
429.4 crore = 4 Billion (2^32) 32th generation ancestors - yr 1055 CE.
858.8 crore = 8 Billion (2^33) 33th generation ancestors - yr 1025 CE.
1717.6 crore = 16 Billion (2^34) 34th generation ancestors - yr 995 CE.
3435.2 crore = 32 Billion (2^35) 35th generation ancestors - yr 965 CE.
6870.4 crore = 64 Billion (2^36) 36th generation ancestors - yr 935 CE.
13740.8 crore = 128 Billion (2^37) 37th generation ancestors - yr 905 CE.
27481.6 crore = 256 Billion (2^38) 38th generation ancestors - yr 875 CE.
54963.2 crore = 512 Billion (2^39) 39th generation ancestors - yr 845 CE. 500 Billion
109926.4 crore = 1024 Billion (2^40) 40th generation ancestors - yr 815 CE. 1000 Billion
In 815 CE, 1000 Billion ancestors of a single person in 2015 CE. And it keeps doubling because every person has 2 parents and each of those parents has 2 more parents and so on. The numbers are so huge that I don't know how it adds up. To give an idea, the present population of the world is 7 Billion. It seems as If I am missing something but I don't know what I am missing. If I am making some mistake please point out.
Broadly, it seems that the number of ancestors would be too large for even a single person as we go back in time and there seems to a total mixing making everyone relative of each other. So, it seems funny that people talk about AIT or something else based on these markers which become less probabilistic and totally mixed up as we go back in time. Only one ancestor can be found with certainty based on Y chromosome using DNA testing. Mitochondria is passed by every female ancestor, so given the huge number of ancestors , one can be sure that all sorts of Mitochondria will be there.
-
member_28656
- BRFite -Trainee
- Posts: 9
- Joined: 11 Aug 2016 06:14
Re: Out-of-India - From Theory to Truth: Part 2
https://en.wikipedia.org/wiki/Genealogical_DNA_test
Hi johneeG,
Have a look at this Wiki page. DNA testing is a mix of something called STR (Short tandem repeat) and SNP (Single nucleotide polymorphism) testing, in both Y-chromosome and Mitochondrial DNA. You are right in saying that the results are more statistical and combinatorial.
In SNP testing the average rate at which DNA sequence changes de novo (i.e. on its own/naturally) is known and so, you can count the number and type of changes that have happened between an ancestor and descendant and the distance between them both in terms of time/generations and mixing with other groups is estimated. Similar deal with STR.
I am a molecular biologist (not a population geneticist), my understanding of this specific field is limited.
Hi johneeG,
Have a look at this Wiki page. DNA testing is a mix of something called STR (Short tandem repeat) and SNP (Single nucleotide polymorphism) testing, in both Y-chromosome and Mitochondrial DNA. You are right in saying that the results are more statistical and combinatorial.
In SNP testing the average rate at which DNA sequence changes de novo (i.e. on its own/naturally) is known and so, you can count the number and type of changes that have happened between an ancestor and descendant and the distance between them both in terms of time/generations and mixing with other groups is estimated. Similar deal with STR.
I am a molecular biologist (not a population geneticist), my understanding of this specific field is limited.
Re: Out-of-India - From Theory to Truth: Part 2
RaviK saar,RaviK wrote:https://en.wikipedia.org/wiki/Genealogical_DNA_test
Hi johneeG,
Have a look at this Wiki page. DNA testing is a mix of something called STR (Short tandem repeat) and SNP (Single nucleotide polymorphism) testing, in both Y-chromosome and Mitochondrial DNA. You are right in saying that the results are more statistical and combinatorial.
In SNP testing the average rate at which DNA sequence changes de novo (i.e. on its own/naturally) is known and so, you can count the number and type of changes that have happened between an ancestor and descendant and the distance between them both in terms of time/generations and mixing with other groups is estimated. Similar deal with STR.
I am a molecular biologist (not a population geneticist), my understanding of this specific field is limited.
thanks for replying.
How is this STR & SNP connected to Y-Chromosome and Mitochondria? And what is autosome?
Another thing I can't understand is the the number of ancestors is rising with a geometric progression of 2 at each stage. And if it continues like that, then it is around 1000 Billion within 1000 years(from 2000 CE). How can it be when the world population today is supposed to be just 7 Billion? What am I missing?
-
member_28656
- BRFite -Trainee
- Posts: 9
- Joined: 11 Aug 2016 06:14
Re: Out-of-India - From Theory to Truth: Part 2
No mention johneeG Saar 
Every human cell has 46 chromosomes or 23 pairs of chromosomes, out of which, 22 pairs are regular ones (autosomes) and the last pair sex-chromosomes (Allosomes, either XX as a pair or XY as a pair).
To put it in easy terms, DNA is made up of 4 nucleotide bases (A, T, G and C). A always binds to T and G always binds to C. These are base pairs. so for example, at a particular locus (address) on any chromosome (can be any, including Y), the sequence will read something like: ATGCATGCATGC. If there is a SNP/mutation, this sequence might read ATGCA"C"GCATGC, meaning there is a T>C change. Based on statistical and chemical modelling, rate of these changes can be estimated (https://en.wikipedia.org/wiki/Mutation_rate).
SNPs occur all over the Genome (Entire DNA of a cell). some SNPs are so specific to certain population groups, they are called founder mutations. (https://en.wikipedia.org/wiki/Founder_effect). So, SNPs also occur on Y-Chromosome and mitochondrial DNA.
STR (Short tandem repeats) or microsatellite sequences are non-coding sequences. They are repetitive sequences ex: TATATATATA or GTCGTCGTCGTCGTC . They can exist in tandem. i.e. in one locus, for a trinucleotide STR, GTCGTCGTCGTCGTC, GTC is repeated and it can repeat for up to 50 times. This is specific like a barcode, thats why they are used in DNA finger printing/forensics. so in say, 50 loci, 50 STRs, can repeat for different number of times. Again STRs/microsatellites occur on every chromosome.
WRT number of ancestors, till recently it was normal for a couple to have 10-15 kids. So it might be a reverse funnel, each generation giving rise to more offsprings than parents. A couple could give rise to 10 kids. So, untill say 1950, every generation was larger than the one that preceded it. Apart from that, the natural adjustments like earthquakes, epidemics, wars, volcanic eruptions also helped keep the population in check. If you take this into account, the number of ancestors would be much smaller.
Every human cell has 46 chromosomes or 23 pairs of chromosomes, out of which, 22 pairs are regular ones (autosomes) and the last pair sex-chromosomes (Allosomes, either XX as a pair or XY as a pair).
To put it in easy terms, DNA is made up of 4 nucleotide bases (A, T, G and C). A always binds to T and G always binds to C. These are base pairs. so for example, at a particular locus (address) on any chromosome (can be any, including Y), the sequence will read something like: ATGCATGCATGC. If there is a SNP/mutation, this sequence might read ATGCA"C"GCATGC, meaning there is a T>C change. Based on statistical and chemical modelling, rate of these changes can be estimated (https://en.wikipedia.org/wiki/Mutation_rate).
SNPs occur all over the Genome (Entire DNA of a cell). some SNPs are so specific to certain population groups, they are called founder mutations. (https://en.wikipedia.org/wiki/Founder_effect). So, SNPs also occur on Y-Chromosome and mitochondrial DNA.
STR (Short tandem repeats) or microsatellite sequences are non-coding sequences. They are repetitive sequences ex: TATATATATA or GTCGTCGTCGTCGTC . They can exist in tandem. i.e. in one locus, for a trinucleotide STR, GTCGTCGTCGTCGTC, GTC is repeated and it can repeat for up to 50 times. This is specific like a barcode, thats why they are used in DNA finger printing/forensics. so in say, 50 loci, 50 STRs, can repeat for different number of times. Again STRs/microsatellites occur on every chromosome.
WRT number of ancestors, till recently it was normal for a couple to have 10-15 kids. So it might be a reverse funnel, each generation giving rise to more offsprings than parents. A couple could give rise to 10 kids. So, untill say 1950, every generation was larger than the one that preceded it. Apart from that, the natural adjustments like earthquakes, epidemics, wars, volcanic eruptions also helped keep the population in check. If you take this into account, the number of ancestors would be much smaller.
Re: Out-of-India - From Theory to Truth: Part 2
RaviK wrote:Every human cell has 46 chromosomes or 23 pairs of chromosomes, out of which, 22 pairs are regular ones (autosomes) and the last pair sex-chromosomes (Allosomes, either XX as a pair or XY as a pair).
To put it in easy terms, DNA is made up of 4 nucleotide bases (A, T, G and C). A always binds to T and G always binds to C. These are base pairs. so for example, at a particular locus (address) on any chromosome (can be any, including Y), the sequence will read something like: ATGCATGCATGC. If there is a SNP/mutation, this sequence might read ATGCA"C"GCATGC, meaning there is a T>C change. Based on statistical and chemical modelling, rate of these changes can be estimated (https://en.wikipedia.org/wiki/Mutation_rate).
SNPs occur all over the Genome (Entire DNA of a cell). some SNPs are so specific to certain population groups, they are called founder mutations. (https://en.wikipedia.org/wiki/Founder_effect). So, SNPs also occur on Y-Chromosome and mitochondrial DNA.
STR (Short tandem repeats) or microsatellite sequences are non-coding sequences. They are repetitive sequences ex: TATATATATA or GTCGTCGTCGTCGTC . They can exist in tandem. i.e. in one locus, for a trinucleotide STR, GTCGTCGTCGTCGTC, GTC is repeated and it can repeat for up to 50 times. This is specific like a barcode. so in say, 50 loci, 50 STRs, can repeat for different number of times. Again STRs/microsatellites occur on every chromosome.
WRT number of ancestors, till recently it was normal for a couple to have 10-15 kids. So it might be a reverse funnel, each generation giving rise to more offsprings than parents. A couple could give rise to 10 kids. So, untill say 1950, every generation was larger than the one that preceded it. Apart from that, the natural adjustments like earthquakes, epidemics, wars, volcanic eruptions also helped keep the population in check. If you take this into account, the number of ancestors would be much smaller.
Thanks RaviK Saar.
Just found an article which says the same thing.
LinkSTRs vs SNPs, Multiple DNA Personalities
Posted on February 10, 2014
One of the questions I receive rather regularly is about the difference between STRs and SNPs.
Generally, what people really want to understand is the difference between the products, and a basic answer is really all they want. I explain that an STR or Short Tandem Repeat is a different kind of a mutation than a SNP or a Single Nucleotide Polymorphism. STRs are useful genealogically, to determine to whom you match within a recent timeframe, of say, the past 500 years or so, and SNPs define haplogroups which reach much further back in time. Furthermore SNPs are considered “once in a lifetime,” or maybe better stated, “once in the lifetime of mankind” type of events, known as a UEP, Unique Event Polymorphism, where STRs happen “all the time,” in every haplogroup. In fact, this is why you can check for the same STR markers in every haplogroup – those markers we all know and love.
This was a pretty good explanation for a long time but as sequencing technology has improved and new tests have become available, such as the Full Y and Big Y tests, new mutations are being very rapidly discovered which blurs the line between the timeframes that had been used to separate these types of tests. In fact, now they are overlapping in time, so SNPs are, in some cases becoming genealogically useful. This also means that these newly discovered family SNPs are relatively new, meaning they only occurred between the current generation and 1000 years ago, so we should not expect to find huge numbers of these newly developed mutations in the population. For example, if the SNP that defined haplogroup R1b1a2, M269, occurred 15,000 years ago in one man, his descendants have had 15,000 years to procreate and pass his M269 on down the line(s), something they have done very successfully since about half of Europe is either M269 or a subclade.
Each subclade has a SNP all its own. In fact, each subclade is defined by a specific SNP that forms its own branch of the human Y haplotree.
So far, so good.
But what does a SNP or an STR really look like, I mean, in the raw data? How do you know that you’re seeing one or the other?
Like Baseball – 4 Bases
The smallest units of DNA are made up of 4 base nucleotides, DNA words, that are represented by the following letters:
A = Adenine
C = Cytosine
G = Guanine
T = Thymine
These nucleotides combine in pairs to form the ladder rungs of DNA, shown right that connect the helix backbones. T typically combines with A and C usually combines with G, reaching between the backbones of the double helix to connect with their companion protein in the center.
You don’t need to remember the words or even the letters, just remember that we are looking for pattern matches of segments of DNA.
Point Mutations
Your DNA when represented on paper looks like a string of beads where there are 4 kinds of beads, each representing one of the nucleotides above. One segment of your DNA might look like this:
If this is what the standard or reference sequence for your haplotype (your personal DNA results) or your family haplogroup (ancestral clan) looks like, then a mutation would be defined as any change, addition, or deletion. A change would be if the first A above were to change to T or G or C as in the example below:
A deletion would be noticed if the leading A were simply gone.
An addition of course would be if a new bead were inserted in the sequence at that location.
All of the above changes involve only one location. These are all known as Point Mutations, because they occur at one single point.
SNPs
A point mutation may or may not be a SNP. A SNP is defined by geneticists as a point mutation that is found in more than 1% of the population. This should tell you right away that when we say “we’ve discovered a new SNP,” we’re really mis-applying that term, because until we determine that the frequency which it is found in the population is over the 1% threshold, it really isn’t a SNP, but is still considered a point mutation or binary polymorphism.
Today, when SNPS, or point mutations are discovered, they are considered “private mutations” or “family mutations.” There has been consternation for some time about how to handle these types of situations. ISOGG has set forth their criteria on their website. They currently have the most comprehensive tree, but they certainly have their work cut out for them with the incoming tsunami of new SNPS that will be discovered utilizing these next generation tests, hundreds of which are currently in process.
STRs
A STR, or Short Tandem Repeat is analogous to a genetic stutter, or the copy machine getting stuck. In the same situation as above, utilizing the same base for comparison, we see a group of inserted nucleotides that are all duplicates of each other.
In this case, we have a short tandem repeat that is 4 segments in length meaning that CT is inserted 4 times. To translate, if this is marker DYS marker 390, you have a value of 5, meaning 5 repeats of CT.
So I’ve been fat and happy with this now for years, well over a decade.
The Monkey Wrench
And then I saw this:
“The L69/L159 polymorphism is essentially a SNP/STR oxymoron.”
To the best of my knowledge, this is impossible – one type of mutation excludes the other. I googled about this topic and found nothing, nor did I find additional discussion of L69, other than this.
My first reaction to this was “that’s impossible,” followed by “Bloody Hell,” and my next reaction was to find someone who knew.
I reached out to Dr. David Mittelman, geneticist and Chief Scientific Officer at Gene by Gene, parent company of Family Tree DNA. I asked him about the SNP/STR oxymoron and he said:
“This is impossible. There is no such thing as a SNP/STR.”
Whew! I must say, I’m relieved. I thought there for a minute there I had lost my mind.
I asked him what is really going on in this sequence, and he replied that, “This would be a complex variant — when multiple things are happening at once.”
Now, that I understand. I have children, and grandchildren – I fully understand multiple things happening at once. Let’s break this example apart and take a look at what is really happening.
HUGO is a reference standard, so let’s start there as our basis for comparison.
In the L69 variant we have the following sequence.
HUGO variant 2
We see two distinct things happening in this sequence. First, we have the deletion of two Gs, and secondly, we have the insertion of one additional TG. According to Dr. Mittelman, both of these events are STRs, multiple insertions or deletions, and neither are point mutations or SNPs, so neither of these should really have SNP names, they should have STR type of names.
Let’s look at the L159 variant.
HUGO variant 3
In this case, we have the GG insertion and then we have a TG deletion.
In both cases, L69 and L159, the actual length of the DNA sequence remains the same as the reference, but the contents are different. Both had 2 nucleotides removed and 2 added.
The good news is, as a consumer, that you don’t really need to know this, not at this level. The even better news is that with the new discoveries forthcoming, whether they be STRs or SNPs, at the leafy end of the branch, they are often now overlapping with SNPs becoming much more genealogically useful. In the past, if you were looking at a genetics mutation timeline, you had STRs that covered current to 1000 years, then nothing, then beginning at 5,000 or 10,000 years, you have SNPs that were haplogroup defining.
That gap has been steadily shrinking, and today, there often is no gap, the chasm is gone, and we’re discovering freshly hatched recently-occurring SNPs on a daily basis.
The day is fast approaching when you’ll want the full Y sequence, not to further define your haplogroup, but to further delineate your genealogy lines. You’ll have two tools to do that, SNPs and STRs both, not just one.
I want to thank Dr. Mittelman for his generous assistance with this article.
RaviK saar,
my doubts are:
How is a haplogroup determined?
Also, it needs just a small amount of mixing of populations for this mechanism to go haywire because it is based on small mutations. I think such mutations would be quite common. In recent history, perhaps, probability can be used to make something out of this data. But, its seems really less useful as we go in time more than 1000 years.
About the ancestors and the increasing number:
I think it only makes sense if people are marrying and inter-marrying. Most probably people are marrying cousins(first removed to 4th removed). For example, in MB story, Kunthi is the sister of Krushna's father. Kunthi is the aunt of Krushna. Krushna's sister Subhadhra married Arjuna who is the son of Kunthi. So, Arjuna and Subhadhra are cousins and yet married. I think most of the marriages were of this kind.
Another system would be to give daughter to a family and in return obtained a daughter-in-law from the same family.
Even then, there would be too much mixing because the number of descendents would be large who would marry all over the place. So, overall, I think most of these people married and mixed together and are related to each other. So, in a large timeframe, I think these probabilistic models won't work based on small mutations. These methods may work in a small time-frame of 500-1000 years if endogamy is assumed. But, endogamy is easy to assume and difficult to implement in any society at any time-frame. Thats why we have so many Jaathis which keep increasing in number and keep mutating.
Re: Out-of-India - From Theory to Truth: Part 2
Then an 'open letter to clown xyz' ought to be done. Because the site where he wrote is a one sided motor mouth.Pulikeshi wrote:^^^the article is important as it exposes Mohan Guruswamy's shoddy understanding of history!
This man is so worried about ultra-nationalism that he is willing to ignore the exta-nationals invading the very identity of Indians.
The issue can be ignored at great peril given educated folks like Kharge and Gurusswamy endorse the AIT nonsense.
I'll try to dissect whatever I could in tweets.
-
member_28656
- BRFite -Trainee
- Posts: 9
- Joined: 11 Aug 2016 06:14
Re: Out-of-India - From Theory to Truth: Part 2
The high end statistical modelling and Bayesian mathematics go over my headjohneeG wrote: RaviK saar,
my doubts are:
How is a haplogroup determined?
Also, it needs just a small amount of mixing of populations for this mechanism to go haywire because it is based on small mutations. I think such mutations would be quite common. In recent history, perhaps, probability can be used to make something out of this data. But, its seems really less useful as we go in time more than 1000 years.
About the ancestors and the increasing number:
I think it only makes sense if people are marrying and inter-marrying. Most probably people are marrying cousins(first removed to 4th removed). For example, in MB story, Kunthi is the sister of Krushna's father. Kunthi is the aunt of Krushna. Krushna's sister Subhadhra married Arjuna who is the son of Kunthi. So, Arjuna and Subhadhra are cousins and yet married. I think most of the marriages were of this kind.
I am good with the technology part since I use the same methods for my job but application is different (cancer genomics).
From my understanding, after a while of living with limited breeding with other groups, each population develops a cluster of SNPs and STRs that are some what unique to that population, this unique pattern is a Haplotype. We are not talking about a small amount of time but more like hundreds of years if not more. And, the number of Loci which are analysed for these SNPs and STRs is huge. Some studies even test tens of thousands. Larger the number of Loci studied, more robust your estimations are. IMHO, most forensic labs that do this kind of testing just do the bare minimum number to serve the purpose(i.e. identification of people) at a minimum cost while population based genetic studies are funded to study a large number of these loci.
These papers should give you a better idea.
Inference of Population Structure Using Multilocus Genotype Data (http://www.genetics.org/content/155/2/945.long)
Detecting immigration by using multilocus genotypes (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC23111/)
And of course there is always wiki
-
member_28656
- BRFite -Trainee
- Posts: 9
- Joined: 11 Aug 2016 06:14
Re: Out-of-India - From Theory to Truth: Part 2
So more isolated a group is, more unique their genetic profile would be, albeit with limited diversity. The group size (and by extension their geographical spread) would also have an effect. And in theory there should be a tipping point where if the genetic diversity falls below that, that group would disintegrate by natural selection. Lucky us, we dont marry within our Gothras (ensuring a minimum level of divergence)
Re: Out-of-India - From Theory to Truth: Part 2
https://www.youtube.com/watch?v=xCY2lJuke_M
Hinduism in Russia - a vignette
https://www.youtube.com/watch?v=-QJJzLc8bbk
Hindu temple discovered (old, but may not be here).
Hinduism in Russia - a vignette
https://www.youtube.com/watch?v=-QJJzLc8bbk
Hindu temple discovered (old, but may not be here).
Re: Out-of-India - From Theory to Truth: Part 2
From Twitter:
manasataramgini
@blog_supplement
1 can estimate that approximately at least 50% of the Y-chromosomes of varNa populations as a group originated from invading Aryan males
Re: Out-of-India - From Theory to Truth: Part 2
http://www.sci-news.com/paleontology/sc ... 02293.html
55-Million-Year-Old Fossils Suggest Ancestor of Rhinos, Horses Originated in India
55-Million-Year-Old Fossils Suggest Ancestor of Rhinos, Horses Originated in India
Re: Out-of-India - From Theory to Truth: Part 2
he is another sarkari leftist. i used to follow him on FB. another elite chap who claims to be aware of everything and anything. any brfite could take his arguments to shreds.Pulikeshi wrote:^^^the article is important as it exposes Mohan Guruswamy's shoddy understanding of history!
This man is so worried about ultra-nationalism that he is willing to ignore the exta-nationals invading the very identity of Indians.
The issue can be ignored at great peril given educated folks like Kharge and Gurusswamy endorse the AIT nonsense.
Re: Out-of-India - From Theory to Truth: Part 2
So, he is saying that upper castes are progeny of foreign invaders? I think this is nonsense. Firstly, today, India's population is much bigger than anything in Europe or central Asia. India's population: 1.2 Billion. Europe Population 742 million. Central Asian Population 66 million. Even the combined population of Europe and central Asia would not be equal to India. If there is genetic similarity, then it would be reasonable to assume that the larger group is the ancestor and the smaller group is the progeny. I really don't know how he 'estimates' it to be 'invaders'. And if it is invaders, then why would only the 'upper castes' have the genetic similarity? The only way this would make sense is if the 'upper castes' went out of India and developed genetic relations in Europe or Central Asia. Anyway, I have my doubts on this whole field. I think everyone is connected to everyone. Based on 0.1% deviation, they are trying to make huge estimations going thousands years back based on small samples(compared to the whole population).A_Gupta wrote:From Twitter:manasataramgini
@blog_supplement
1 can estimate that approximately at least 50% of the Y-chromosomes of varNa populations as a group originated from invading Aryan males
Re: Out-of-India - From Theory to Truth: Part 2
Utter BullshitA_Gupta wrote:From Twitter:manasataramgini
@blog_supplement
1 can estimate that approximately at least 50% of the Y-chromosomes of varNa populations as a group originated from invading Aryan males
Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CE et al. (2006) Polarity and tem-
porality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous
expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 78:202–221.
Although considerable cultural impact on social hierarchy and language in South Asia is attributable to the arrival
of nomadic Central Asian pastoralists, genetic data (mitochondrial and Y chromosomal) have yielded dramatically
conflicting inferences on the genetic origins of tribes and castes of South Asia. We sought to resolve this conflict,
using high-resolution data on 69 informative Y-chromosome binary markers and 10 microsatellite markers from
a large set of geographically, socially, and linguistically representative ethnic groups of South Asia. We found that
the influence of Central Asia on the pre-existing gene pool was minor. The ages of accumulated microsatellite
variation in the majority of Indian haplogroups exceed 10,000–15,000 years, which attests to the antiquity of
regional differentiation. Therefore, our data do not support models that invoke a pronounced recent genetic input
from Central Asia to explain the observed genetic variation in South Asia. R1a1 and R2 haplogroups indicate
demographic complexity that is inconsistent with a recent single history. Associated microsatellite analyses of the
high-frequency R1a1 haplogroup chromosomes indicate independent recent histories of the Indus Valley and the
peninsular Indian region. Our data are also more consistent with a peninsular origin of Dravidian speakers than
a source with proximity to the Indus and with significant genetic input resulting from demic diffusion associated
with agriculture. Our results underscore the importance of marker ascertainment for distinguishing phylogenetic
terminal branches from basal nodes when attributing ancestral composition and temporality to either indigenous
or exogenous sources. Our reappraisal indicates that pre-Holocene and Holocene-era—not Indo-European—ex-
pansions have shaped the distinctive South Asian Y-chromosome landscape.
Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J et al. (2003) The genetic heritage of the
earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72: 313–332.
Two tribal groups from southern India—the Chenchus and Koyas—were analyzed for variation in mitochondrial
DNA (mtDNA), the Y chromosome, and one autosomal locus and were compared with six caste groups from
different parts of India, as well as with western and central Asians. In mtDNA phylogenetic analyses, the Chenchus
and Koyas coalesce at Indian-specific branches of haplogroups M and N that cover populations of different social
rank from all over the subcontinent. Coalescence times suggest early late Pleistocene settlement of southern Asia
and suggest that there has not been total replacement of these settlers by later migrations. H, L, and R2 are the
major Indian Y-chromosomal haplogroups that occur both in castes and in tribal populations and are rarely found
outside the subcontinent. Haplogroup R1a, previously associated with the putative Indo-Aryan invasion, was found
at its highest frequency in Punjab but also at a relatively high frequency (26%) in the Chenchu tribe. This finding,
together with the higher R1a-associated short tandem repeat diversity in India and Iran compared with Europe
and central Asia, suggests that southern and western Asia might be the source of this haplogroup. Haplotype frequencies of the MX1 locus of chromosome 21 distinguish Koyas and Chenchus, along with Indian caste groups, from European and eastern Asian populations. Taken together, these results show that Indian tribal and caste populations derive largely from the same genetic heritage of Pleistocene southern and western Asians and have received limited gene flow from external regions since the Holocene. The phylogeography of the primal mtDNAand Y-chromosome founders suggests that these southern Asian Pleistocene coastal settlers from Africa would have provided the inocula for the subsequent differentiation of the distinctive eastern and western Eurasian gene pools
Re: Out-of-India - From Theory to Truth: Part 2
Having knowledge, analytical skills and way of articulation is one thing and can be humbly admitted in anybody, regardless of their position.
But it is a folly to build over confidence and start thinking 'Only I know', 'I know all', 'I am the only authentic torchbearer of bla bla bla'.
He is critical of BR, Swarajya magazine and many others (perhaps not India Facts, his articles show up there) saying that these aren't actually Hindu friendly forces and have been infiltrated and what not.
Other than these antics and AIT bullshit, I've found his posts on medieval history very useful.
But it is a folly to build over confidence and start thinking 'Only I know', 'I know all', 'I am the only authentic torchbearer of bla bla bla'.
He is critical of BR, Swarajya magazine and many others (perhaps not India Facts, his articles show up there) saying that these aren't actually Hindu friendly forces and have been infiltrated and what not.
Other than these antics and AIT bullshit, I've found his posts on medieval history very useful.
Re: Out-of-India - From Theory to Truth: Part 2
shiv wrote:Utter BullshitA_Gupta wrote:From Twitter:
manasataramgini
@blog_supplement
1 can estimate that approximately at least 50% of the Y-chromosomes of varNa populations as a group originated from invading Aryan males
Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CE et al. (2006) Polarity and tem-
porality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous
expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 78:202–221.
Although considerable cultural impact on social hierarchy and language in South Asia is attributable to the arrival
of nomadic Central Asian pastoralists, genetic data (mitochondrial and Y chromosomal) have yielded dramatically
conflicting inferences on the genetic origins of tribes and castes of South Asia. We sought to resolve this conflict,
using high-resolution data on 69 informative Y-chromosome binary markers and 10 microsatellite markers from
a large set of geographically, socially, and linguistically representative ethnic groups of South Asia. We found that
the influence of Central Asia on the pre-existing gene pool was minor. The ages of accumulated microsatellite
variation in the majority of Indian haplogroups exceed 10,000–15,000 years, which attests to the antiquity of
regional differentiation. Therefore, our data do not support models that invoke a pronounced recent genetic input
from Central Asia to explain the observed genetic variation in South Asia. R1a1 and R2 haplogroups indicate
demographic complexity that is inconsistent with a recent single history. Associated microsatellite analyses of the
high-frequency R1a1 haplogroup chromosomes indicate independent recent histories of the Indus Valley and the
peninsular Indian region. Our data are also more consistent with a peninsular origin of Dravidian speakers than
a source with proximity to the Indus and with significant genetic input resulting from demic diffusion associated
with agriculture. Our results underscore the importance of marker ascertainment for distinguishing phylogenetic
terminal branches from basal nodes when attributing ancestral composition and temporality to either indigenous
or exogenous sources. Our reappraisal indicates that pre-Holocene and Holocene-era—not Indo-European—ex-
pansions have shaped the distinctive South Asian Y-chromosome landscape.
The 2015 paper by Underhill et. al. suggests that India can not be considered the origin of R1a.Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J et al. (2003) The genetic heritage of the
earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72: 313–332.
Two tribal groups from southern India—the Chenchus and Koyas—were analyzed for variation in mitochondrial
DNA (mtDNA), the Y chromosome, and one autosomal locus and were compared with six caste groups from
different parts of India, as well as with western and central Asians. In mtDNA phylogenetic analyses, the Chenchus
and Koyas coalesce at Indian-specific branches of haplogroups M and N that cover populations of different social
rank from all over the subcontinent. Coalescence times suggest early late Pleistocene settlement of southern Asia
and suggest that there has not been total replacement of these settlers by later migrations. H, L, and R2 are the
major Indian Y-chromosomal haplogroups that occur both in castes and in tribal populations and are rarely found
outside the subcontinent. Haplogroup R1a, previously associated with the putative Indo-Aryan invasion, was found
at its highest frequency in Punjab but also at a relatively high frequency (26%) in the Chenchu tribe. This finding,
together with the higher R1a-associated short tandem repeat diversity in India and Iran compared with Europe
and central Asia, suggests that southern and western Asia might be the source of this haplogroup. Haplotype frequencies of the MX1 locus of chromosome 21 distinguish Koyas and Chenchus, along with Indian caste groups, from European and eastern Asian populations. Taken together, these results show that Indian tribal and caste populations derive largely from the same genetic heritage of Pleistocene southern and western Asians and have received limited gene flow from external regions since the Holocene. The phylogeography of the primal mtDNAand Y-chromosome founders suggests that these southern Asian Pleistocene coastal settlers from Africa would have provided the inocula for the subsequent differentiation of the distinctive eastern and western Eurasian gene pools
Re: Out-of-India - From Theory to Truth: Part 2
Does he say what he bases his estimation on?A_Gupta wrote:From Twitter:manasataramgini
@blog_supplement
1 can estimate that approximately at least 50% of the Y-chromosomes of varNa populations as a group originated from invading Aryan males
Re: Out-of-India - From Theory to Truth: Part 2
A_Gupta wrote:From Twitter:manasataramgini
@blog_supplement
1 can estimate that approximately at least 50% of the Y-chromosomes of varNa populations as a group originated from invading Aryan males
But bigger population ain't a clincher. Cheetahs (from chitkabra) got their names in India but we don't have them in India anymore.johneeG wrote: So, he is saying that upper castes are progeny of foreign invaders? I think this is nonsense. Firstly, today, India's population is much bigger than anything in Europe or central Asia. India's population: 1.2 Billion. Europe Population 742 million. Central Asian Population 66 million. Even the combined population of Europe and central Asia would not be equal to India. If there is genetic similarity, then it would be reasonable to assume that the larger group is the ancestor and the smaller group is the progeny. I really don't know how he 'estimates' it to be 'invaders'. And if it is invaders, then why would only the 'upper castes' have the genetic similarity? The only way this would make sense is if the 'upper castes' went out of India and developed genetic relations in Europe or Central Asia. Anyway, I have my doubts on this whole field. I think everyone is connected to everyone. Based on 0.1% deviation, they are trying to make huge estimations going thousands years back based on small samples(compared to the whole population).
Re: Out-of-India - From Theory to Truth: Part 2
http://www.nature.com/ejhg/journal/v23/ ... 1450a.html
2014 paper by Underhill, etc.
2014 paper by Underhill, etc.
Re: Out-of-India - From Theory to Truth: Part 2
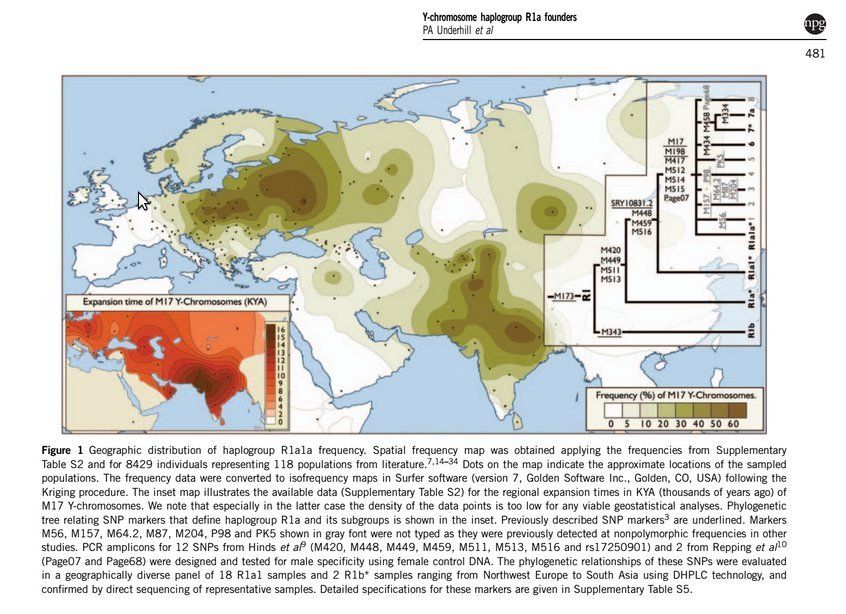
Link please. The 2014 paper posted by AGupta does not say that. The only "South Asia Origin" part of R1a is R1a1a1 (M 17). The Underhill paper posted above goes into more detail down below that and looks at origins of those.peter wrote: The 2015 paper by Underhill et. al. suggests that India can not be considered the origin of R1a.
This image - showing the oldest origins of M17 in India (Dark Brown) is from the 2010 Underhill paper
-
Nilesh Oak
- BRFite
- Posts: 1670
- Joined: 11 Aug 2016 06:14
Re: Out-of-India - From Theory to Truth: Part 2
Virendra ji, what is his name again?Virendra wrote:Having knowledge, analytical skills and way of articulation is one thing and can be humbly admitted in anybody, regardless of their position.
But it is a folly to build over confidence and start thinking 'Only I know', 'I know all', 'I am the only authentic torchbearer of bla bla bla'.
He is critical of BR, Swarajya magazine and many others (perhaps not India Facts, his articles show up there) saying that these aren't actually Hindu friendly forces and have been infiltrated and what not.
Other than these antics and AIT bullshit, I've found his posts on medieval history very useful.
Re: Out-of-India - From Theory to Truth: Part 2
Manasataramgini?Nilesh Oak wrote:Virendra ji, what is his name again?Virendra wrote:Having knowledge, analytical skills and way of articulation is one thing and can be humbly admitted in anybody, regardless of their position.
But it is a folly to build over confidence and start thinking 'Only I know', 'I know all', 'I am the only authentic torchbearer of bla bla bla'.
He is critical of BR, Swarajya magazine and many others (perhaps not India Facts, his articles show up there) saying that these aren't actually Hindu friendly forces and have been infiltrated and what not.
Other than these antics and AIT bullshit, I've found his posts on medieval history very useful.
This is his blog:
https://manasataramgini.wordpress.com/
His name is Arvind, I think.
This is his latest:
https://manasataramgini.wordpress.com/2 ... europeans/
There have been a whole lot of developments in ancient human genomics that have more or less solved key issues pertaining to the early Indo-European expansions. We would like to discuss these but then it would need a long article which we are not currently inclined to write because it would need enormous amount of data to be presented in an understandable form. Moreover, there are indications via various channels that, exciting as the current developments are, there are going to be new ones which will make things even clearer with respect to the Indian situation. Hence, we were somewhat disinclined to engage in any long writing on this topic. Nevertheless, we could not control our temptation to at least say a few words in this regard.
About 21-22 years ago a strange, new aberration in Hindu thought came to our attention: The Out-of-India-theory (OIT), which posited that the Indo-Aryans were autochthons of the Indian subcontinent. At first we brushed it aside as being a mere fantasy of some ill-educated raconteurs, who might simultaneously see Tipoo Sultan as a freedom fighter. But as the 1990s came to an end the the 2000s began this stream of thinking became dominant among the Hindus. So much so that most politically pro-Hindu individuals also tied themselves to some version of OIT. Across different fora you would see them thundering as though they were Parjanya: “The Aryan invasion is a myth.” They started seeing it as an instrument created by the English or more generally the Leukosphere to sow dissension among the autochthonous Hindus. We can provide a long list of prominent Hindus on the internet and associates of Hindus who were proponents of some form OIT: S. Talageri, S. Kak, N.S. Rajaram, V. Agrawal, B.B. Lal, S. Kalyanaraman, D. Frawley, R. Malhotra, M. Danino, K. Elst, N. Kazanas and so on.
etc.In 2004 Cordaux et al published a clear and simple article titled “Independent Origins of Indian Caste and Tribal Paternal Lineages” that put the genie of OIT firmly back in the bottle. Yet, the Hindus including Talageri went on as though nothing had happened. There were some issues with that old article and it was based only on Y-chromosome haplogroups, but the wealth of new data on the genome-scale that has emerged since has only gone on to confirm the AIT and provide several interesting new details. In the past two years a wealth of ancient DNA is literally making the skeletons, if not the pots, of the steppe cultures speak and they are telling us great stories. Much of this work is done from a Eurocentric viewpoint but it has tremendous implications for us because we are the “other branch of Indo-European”. So what has happened?
Re: Out-of-India - From Theory to Truth: Part 2
shiv wrote:Link please. The 2014 paper posted by AGupta does not say that. The only "South Asia Origin" part of R1a is R1a1a1 (M 17). The Underhill paper posted above goes into more detail down below that and looks at origins of those.peter wrote: The 2015 paper by Underhill et. al. suggests that India can not be considered the origin of R1a.
This image - showing the oldest origins of M17 in India (Dark Brown) is from the 2010 Underhill paper

Origin of hg R1a
To infer the geographic origin of hg R1a-M420, we identified populations harboring at least one of the two most basal haplogroups and possessing high haplogroup diversity. Among the 120 populations with sample sizes of at least 50 individuals and with at least 10% occurrence of R1a, just 6 met these criteria, and 5 of these 6 populations reside in modern-day Iran. Haplogroup diversities among the six populations ranged from 0.78 to 0.86 (Supplementary Table 4). Of the 24 R1a-M420*(xSRY10831.2) chromosomes in our data set, 18 were sampled in Iran and 3 were from eastern Turkey. Similarly, five of the six observed R1a1-SRY10831.2*(xM417/Page7) chromosomes were also from Iran, with the sixth occurring in a Kabardin individual from the Caucasus. Owing to the prevalence of basal lineages and the high levels of haplogroup diversities in the region, we find a compelling case for the Middle East, possibly near present-day Iran, as the geographic origin of hg R1a.
From: http://www.nature.com/ejhg/journal/v23/ ... 1450a.html
Re: Out-of-India - From Theory to Truth: Part 2
So bow does cheetah example help in invading aryan theory,especially when Iran could be part of India back then. To say now that Aryans from Iran invaded instead of some TFTA gora Caucasian Aryans from invaded is a bigger farce it looks like, bigger even then AIT. The genetic science approach shows in fact even more glaringly how agendas have been dominating history.
Re: Out-of-India - From Theory to Truth: Part 2
These are the details needed. How many were sampled from India and from where?peter wrote:Among the 120 populations with sample sizes of at least 50 individuals and with at least 10% occurrence of R1a
Re: Out-of-India - From Theory to Truth: Part 2
From the supplementary tables:
Code: Select all
South Asia Ntotal R1a-all
Pakistan North 85 14
Pakistan South 91 30
India North 98 19
India East 124 5
India Northeast 68 19
India Northwest 127 31
India Central 36 5
India South 97 5
Mixed India 40 5
Nepal Tharu 170 14
Nepal Hindu 25 17
India/Hindu/New Delhi 49 17
India/Tribal/Andhra Pradesh 29 8
Re: Out-of-India - From Theory to Truth: Part 2
There is a problem here:A_Gupta wrote:From the supplementary tables:
Code: Select all
South Asia Ntotal R1a-all Pakistan North 85 14 Pakistan South 91 30 India North 98 19 India East 124 5 India Northeast 68 19 India Northwest 127 31 India Central 36 5 India South 97 5 Mixed India 40 5 Nepal Tharu 170 14 Nepal Hindu 25 17 India/Hindu/New Delhi 49 17 India/Tribal/Andhra Pradesh 29 8
The "Last defining Y-chr marker" in most South Asia people is M576. M576 does not even exist in any table or description.
Re: Out-of-India - From Theory to Truth: Part 2
Is M576 above or below M420 on the tree?shiv wrote:There is a problem here:A_Gupta wrote:From the supplementary tables:
Code: Select all
South Asia Ntotal R1a-all Pakistan North 85 14 Pakistan South 91 30 India North 98 19 India East 124 5 India Northeast 68 19 India Northwest 127 31 India Central 36 5 India South 97 5 Mixed India 40 5 Nepal Tharu 170 14 Nepal Hindu 25 17 India/Hindu/New Delhi 49 17 India/Tribal/Andhra Pradesh 29 8
The "Last defining Y-chr marker" in most South Asia people is M576. M576 does not even exist in any table or description.
Re: Out-of-India - From Theory to Truth: Part 2
Bit difficult to understand what you are saying or whom you are asking. Cheetah example I gave to point out that even though I agree with what johnnygee wrote but a smaller population argument may not clinch the debate.vishvak wrote:So bow does cheetah example help in invading aryan theory,especially when Iran could be part of India back then. To say now that Aryans from Iran invaded instead of some TFTA gora Caucasian Aryans from invaded is a bigger farce it looks like, bigger even then AIT. The genetic science approach shows in fact even more glaringly how agendas have been dominating history.
Re: Out-of-India - From Theory to Truth: Part 2
It is not on the tree at all as far as I can tellpeter wrote: Is M576 above or below M420 on the tree?
Another point to note R1 is older than R1a which is older than R1a1 which is older than R1a1a etc.
Even if R1 developed in Iran or Sweden or Timbuktu, it does not rule out a later development of R1a1a1 M17 in India.For example M 458, which comes lower down than M17 apparently developed in Poland/east Europe about 6000 years ago never came to India, but M 17 exists all the way from India to Poland. That means M458 did not come down to India in the last 6000 years
In this particular paper I have been unable to find any reference to the "estimated age" of these genetic markers. Maybe I need to dig into the paper in more detail. Even if R1 developed in Iran unless a movement to India occurred in the last 3000-3500 years AIT is ruled out. Other papers do show that ANI genes occur in Iran etc but admixture occurred more than 10,000 years ago and continued, leading to all Indians having that gene today - even tribals, ruling out the theory of AIT/AMT in the last 3500 years.
The 3500 years date is important for AIT because it is a linguistic date where they say that PIE existed before Sanskrit and Sanskrit came with Aryans 3500 years ago to India (Mitanni 3800 years ago and PIE 4500 years ago is what linguists claim). If they say Sanskrit came with Aryans 10,000 years ago then they will have to produce proof of PIE older than 10,000 years. That is non existent
Last edited by shiv on 20 Dec 2015 07:16, edited 3 times in total.
Re: Out-of-India - From Theory to Truth: Part 2
If a smaller group is connected to a larger group, then it is logical to assume that the smaller group splintered from the larger one. Today, with advent of technology, Europe and central asia are able to support much larger populations. In the past(thousands of years back), the populations of Europe and central asia would be much smaller compared to India.peter wrote:Bit difficult to understand what you are saying or whom you are asking. Cheetah example I gave to point out that even though I agree with what johnnygee wrote but a smaller population argument may not clinch the debate.vishvak wrote:So bow does cheetah example help in invading aryan theory,especially when Iran could be part of India back then. To say now that Aryans from Iran invaded instead of some TFTA gora Caucasian Aryans from invaded is a bigger farce it looks like, bigger even then AIT. The genetic science approach shows in fact even more glaringly how agendas have been dominating history.
Re: Out-of-India - From Theory to Truth: Part 2
In addition, climate changes (ice ages, sea levels, rainfall levels etc) would further dwindle the amount of anthropization a region could support, specially in Europe.